

*Storage: {[sel_prStorage]}
*Shipping: {[sel_prShipping]}
4.5




 *For research use only!
*For research use only!
Change View
| Size | Price | VIP Price | USA Stock *0-1 Day |
Global Stock *5-7 Days |
In Stock | ||
| {[ item.pr_size ]} |
Inquiry
{[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]} {[ getRatePrice(item.pr_usd,item.pr_rate,1,item.pr_is_large_size_no_price, item.discount_usd) ]} {[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]} |
Inquiry {[ getRatePrice(item.pr_usd,item.pr_rate,item.mem_rate,item.pr_is_large_size_no_price, item.vip_usd) ]} | Inquiry {[ item.pr_usastock ]} In Stock Inquiry - | {[ item.pr_chinastock ]} {[ item.pr_remark ]} In Stock 1-2 weeks - Inquiry - | Login | - + | Inquiry |
Please Login or Create an Account to: See VIP prices and availability
1-2weeks
Inquiry
{[ getRatePrice(item.pr_usd,item.pr_rate,item.mem_rate,item.pr_is_large_size_no_price, item.vip_usd) ]}
{[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]}
{[ getRatePrice(item.pr_usd,1,item.mem_rate,item.pr_is_large_size_no_price, item.pr_usd) ]}
Inquiry
{[ getRatePrice(item.pr_usd,item.pr_rate,1,item.pr_is_large_size_no_price, item.vip_usd) ]}
{[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]}
{[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]}
In Stock
- +
Please Login or Create an Account to: See VIP prices and availability
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
| CAS No. : | 94-99-5 |
| Formula : | C7H5Cl3 |
| M.W : | 195.47 |
| MDL No. : | MFCD00000895 |
| InChI Key : | IRSVDHPYXFLLDS-UHFFFAOYSA-N |
| Pubchem ID : | 7212 |
| GHS Pictogram: |

|
| Signal Word: | Danger |
| Hazard Statements: | H314-H290 |
| Precautionary Statements: | P501-P260-P234-P264-P280-P390-P303+P361+P353-P301+P330+P331-P363-P304+P340+P310-P305+P351+P338+P310-P406-P405 |
| Class: | 8 |
| UN#: | 3265 |
| Packing Group: | Ⅱ |
| Num. heavy atoms | 10 |
| Num. arom. heavy atoms | 6 |
| Fraction Csp3 | 0.14 |
| Num. rotatable bonds | 1 |
| Num. H-bond acceptors | 0.0 |
| Num. H-bond donors | 0.0 |
| Molar Refractivity | 46.22 |
| TPSA ? Topological Polar Surface Area: Calculated from |
0.0 ?2 |
| Log Po/w (iLOGP)? iLOGP: in-house physics-based method implemented from |
2.35 |
| Log Po/w (XLOGP3)? XLOGP3: Atomistic and knowledge-based method calculated by |
3.82 |
| Log Po/w (WLOGP)? WLOGP: Atomistic method implemented from |
3.58 |
| Log Po/w (MLOGP)? MLOGP: Topological method implemented from |
4.1 |
| Log Po/w (SILICOS-IT)? SILICOS-IT: Hybrid fragmental/topological method calculated by |
4.08 |
| Consensus Log Po/w? Consensus Log Po/w: Average of all five predictions |
3.58 |
| Log S (ESOL):? ESOL: Topological method implemented from |
-3.84 |
| Solubility | 0.0285 mg/ml ; 0.000146 mol/l |
| Class? Solubility class: Log S scale |
Soluble |
| Log S (Ali)? Ali: Topological method implemented from |
-3.52 |
| Solubility | 0.0597 mg/ml ; 0.000305 mol/l |
| Class? Solubility class: Log S scale |
Soluble |
| Log S (SILICOS-IT)? SILICOS-IT: Fragmental method calculated by |
-4.65 |
| Solubility | 0.00434 mg/ml ; 0.0000222 mol/l |
| Class? Solubility class: Log S scale |
Moderately soluble |
| GI absorption? Gatrointestinal absorption: according to the white of the BOILED-Egg |
Low |
| BBB permeant? BBB permeation: according to the yolk of the BOILED-Egg |
Yes |
| P-gp substrate? P-glycoprotein substrate: SVM model built on 1033 molecules (training set) |
No |
| CYP1A2 inhibitor? Cytochrome P450 1A2 inhibitor: SVM model built on 9145 molecules (training set) |
Yes |
| CYP2C19 inhibitor? Cytochrome P450 2C19 inhibitor: SVM model built on 9272 molecules (training set) |
No |
| CYP2C9 inhibitor? Cytochrome P450 2C9 inhibitor: SVM model built on 5940 molecules (training set) |
No |
| CYP2D6 inhibitor? Cytochrome P450 2D6 inhibitor: SVM model built on 3664 molecules (training set) |
Yes |
| CYP3A4 inhibitor? Cytochrome P450 3A4 inhibitor: SVM model built on 7518 molecules (training set) |
No |
| Log Kp (skin permeation)? Skin permeation: QSPR model implemented from |
-4.78 cm/s |
| Lipinski? Lipinski (Pfizer) filter: implemented from |
0.0 |
| Ghose? Ghose filter: implemented from |
None |
| Veber? Veber (GSK) filter: implemented from |
0.0 |
| Egan? Egan (Pharmacia) filter: implemented from |
0.0 |
| Muegge? Muegge (Bayer) filter: implemented from |
2.0 |
| Bioavailability Score? Abbott Bioavailability Score: Probability of F > 10% in rat |
0.55 |
| PAINS? Pan Assay Interference Structures: implemented from |
0.0 alert |
| Brenk? Structural Alert: implemented from |
1.0 alert: heavy_metal |
| Leadlikeness? Leadlikeness: implemented from |
No; 1 violation:MW<2.0 |
| Synthetic accessibility? Synthetic accessibility score: from 1 (very easy) to 10 (very difficult) |
1.48 |
* All experimental methods are cited from the reference, please refer to the original source for details. We do not guarantee the accuracy of the content in the reference.

| Yield | Reaction Conditions | Operation in experiment |
|---|---|---|
| In isopropyl alcohol; acetone; | EXAMPLE XVI 15 parts of 1-[2,4-dichloro-beta-(2,4-dichlorobenzyloxy)-phenethyl]imidazole and 60 parts of 2,4-dichlorobenzyl chloride are mixed together in 120 parts of acetone as solvent and the resulting mixture is stirred at reflux temperature for 40 hours. At the end of this period, the mixture is warmed to evaporate off most of the solvent and the residue is poured onto diisopropyl ether, whereupon the 1-(2,4-dichlorobenzyl)-3-[2,4-dichloro-beta-(2,4-dichlorobenzyloxy)phenethyl]imidazolium chloride product separates as an oil. The ether is decanted off and the oily residue is washed with warm xylene and thereafter triturated in 2-propanol whereupon the oily product solidified. The solid product is filtered off, washed with 2-propanol and dried to obtain a purified product, 1-(2,4-dichlorobenzyl)-3-[2,4-dichloro-beta-(2,4-dichlorobenzyloxy)phenethyl]-imidazolium chloride, mp. 178.9° C. |
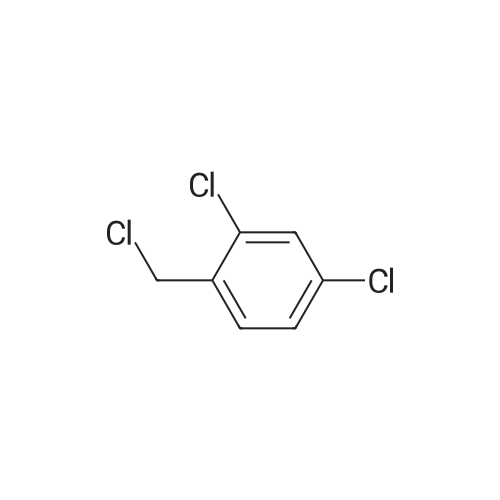
 [ 94-99-5 ]
[ 94-99-5 ]
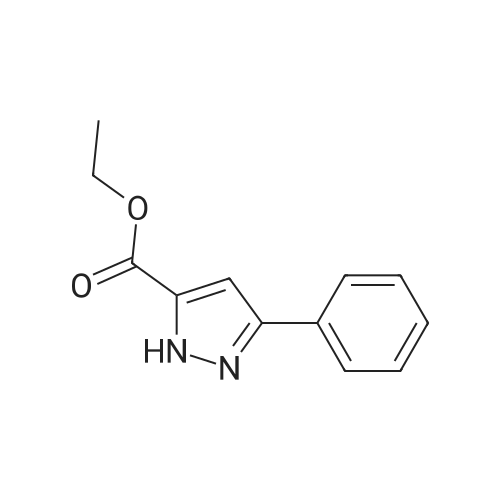 [ 13599-12-7 ]
[ 13599-12-7 ]
| Yield | Reaction Conditions | Operation in experiment |
|---|---|---|
| 78% | With potassium carbonate; In N,N-dimethyl-formamide; at 20℃; | General procedure: To a mixture of 5 (105 g, 573 mmol), K2CO3 (119 g, 860 mmol), and DMF (400 mL) was added benzyl bromide (117 g, 687 mmol). After being stirred at room temperature for 15 h, the mixture was poured into water and extracted with EtOAc. The extract was washed with water and brine, dried over MgSO4, and concentrated under reduced pressure. The residue was purified by silica gel chromatography (hexane/EtOAc, 100:1 to 4:1) to give 6 (130 g, 83percent) as a pale yellow oil. |
| 50% | With potassium carbonate; In N,N-dimethyl-formamide; at 0 - 30℃; for 15h; | A mixture of ethyl 3-phenyl-lH-pyrazole-5-carboxylate (70.0' g) , 2, 4-dichlorobenzyl chloride (69.6 g) , potassium carbonate (53.7 g) and N,N-dimethylformamide (400 ml) was stirred at room temperature for 15 hr, the reaction mixture was concentrated, and the residue was partitioned between water and ethyl acetate. The ethyl acetate layer was washed with water and saturated brine, dried (MgSCU) / and concentrated. The residue was crystallized from ethyl acetate- hexane to give ethyl 1- (2, 4-dichlorobenzyl) -3-phenyl-lH- pyrazole-5-carboxylate (61.0 g, yield: 50percent) as brown crystals, melting point 104-1070C. |
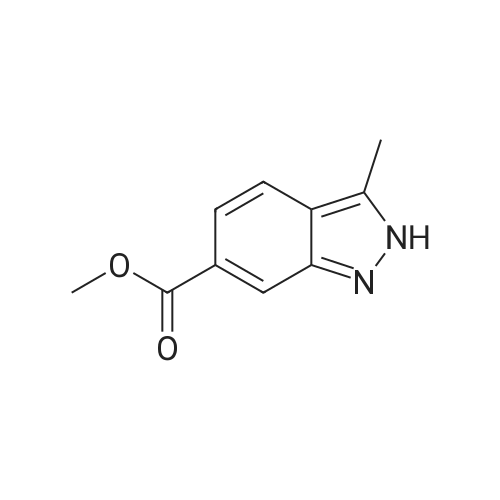 [ 201286-95-5 ]
[ 201286-95-5 ]
 [ 94-99-5 ]
[ 94-99-5 ]
| Yield | Reaction Conditions | Operation in experiment |
|---|---|---|
| 74% | Preparation Example 39-5 1-(2,4-Dichlorobenzyl)-<strong>[201286-95-5]6-(methoxycarbonyl)-3-methyl-1H-indazole</strong> 6-(Methoxycarbonyl)-3-methyl-1H-indazole (0.40 g, 2.1 mmol) was dissolved in dimethylformamide (15 ml) and the mixture was ice-cooled.. sodium hydride (85 mg, 60% suspension in oil, 2.1 mmol as NaH) was added and the mixture was stirred at 0 C. for 30 min. 2,4-Dichlorobenzyl chloride (0.45 g, 2.3 mmol) was added and the mixture was stirred at room temperature for 18 hr.. The reaction mixture was extracted with ethyl acetate/water.. The organic layer was washed with saturated brine and dried over anhydrous sodium sulfate.. The drying agent was filtered off and the filtrate was concentrated under reduced pressure.. The obtained crystalline residue was separated and purified by silica gel column chromatography (eluent: hexane/ethyl acetate=9/1) to give the objective compound (0.54 g, 74%) as colorless crystals. 1H-NMR(CDCl3, delta ppm): 8.06(1H, d, J=1.1 Hz), 7.82(1H, dd, J=1.1 and 8.4 Hz), 7.72(1H, d, J=8.3 Hz), 7.42(1H, d, J=2.0 Hz), 7.08(1H, dd, J=2.0 and 8.3 Hz), 6.60(1H, d, J=8.4 Hz), 5.63(2H, s), 3.94(3H, s), 2.61(3H, s) |
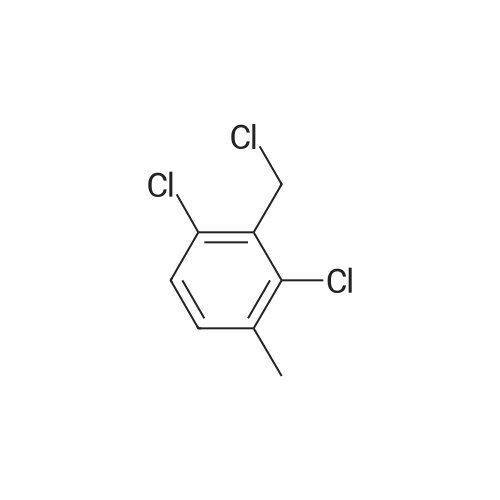
A303298[ 1379325-36-6 ]
1,3-Dichloro-2-(chloromethyl)-4-methylbenzene
Similarity: 0.97
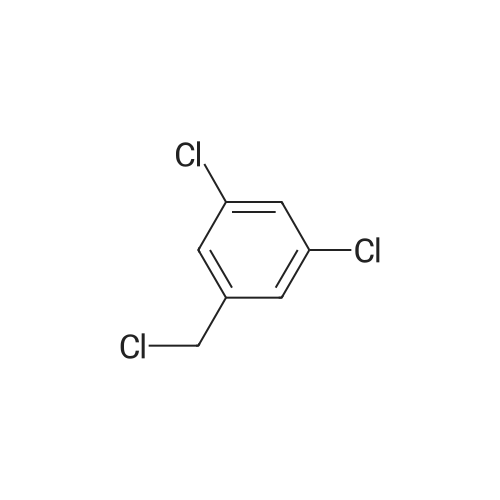
A174518[ 3290-06-0 ]
1,3-Dichloro-5-(chloromethyl)benzene
Similarity: 0.91
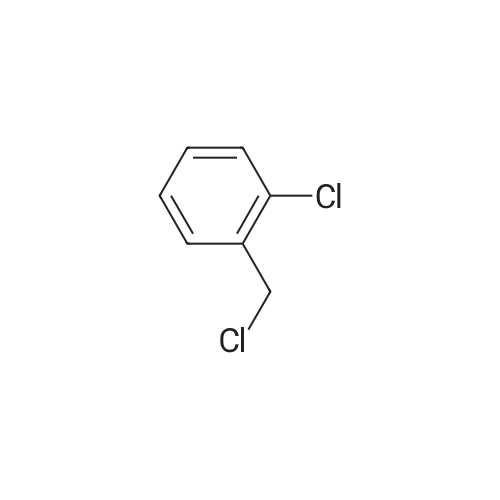
A780623[ 611-19-8 ]
1-Chloro-2-(chloromethyl)benzene
Similarity: 0.91

A303298[ 1379325-36-6 ]
1,3-Dichloro-2-(chloromethyl)-4-methylbenzene
Similarity: 0.97

A174518[ 3290-06-0 ]
1,3-Dichloro-5-(chloromethyl)benzene
Similarity: 0.91

A780623[ 611-19-8 ]
1-Chloro-2-(chloromethyl)benzene
Similarity: 0.91

A303298[ 1379325-36-6 ]
1,3-Dichloro-2-(chloromethyl)-4-methylbenzene
Similarity: 0.97

A174518[ 3290-06-0 ]
1,3-Dichloro-5-(chloromethyl)benzene
Similarity: 0.91

A780623[ 611-19-8 ]
1-Chloro-2-(chloromethyl)benzene
Similarity: 0.91
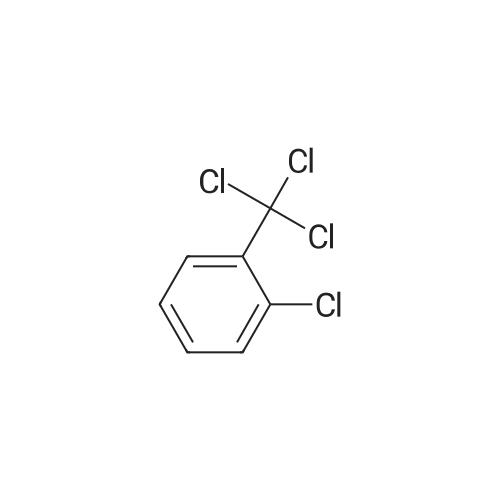
A808560[ 2136-89-2 ]
1-Chloro-2-(trichloromethyl)benzene
Similarity: 0.81
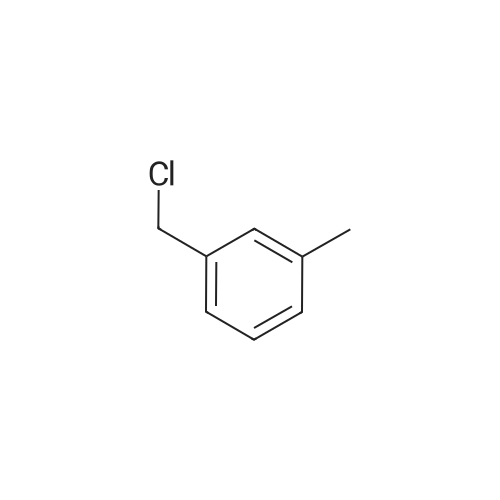
A144534[ 620-19-9 ]
1-(Chloromethyl)-3-methylbenzene
Similarity: 0.71